PARC - phenotyping by accelerated refined community-partitioning
PARC is a fast, automated, combinatorial graph-based clustering approach that integrates hierarchical graph construction (HNSW) and data-driven graph-pruning with the new Leiden community-detection algorithm. Stassen et al. (Bioinformatics, 2020) PARC:ultrafast and accurate clustering of phenotypic data of millions of single cells.
✳️ PARC forms the clustering basis for our new Trajectory Inference (TI) method VIA available on Readthedocs and Github. VIA is a single-cell Trajectory Inference method that offers topology construction and visualization, pseudotimes, automated prediction of terminal cell fates and temporal gene dynamics along detected lineages. VIA can also be used to topologically visualize the graph-based connectivity of clusters found by PARC in a non-TI context.
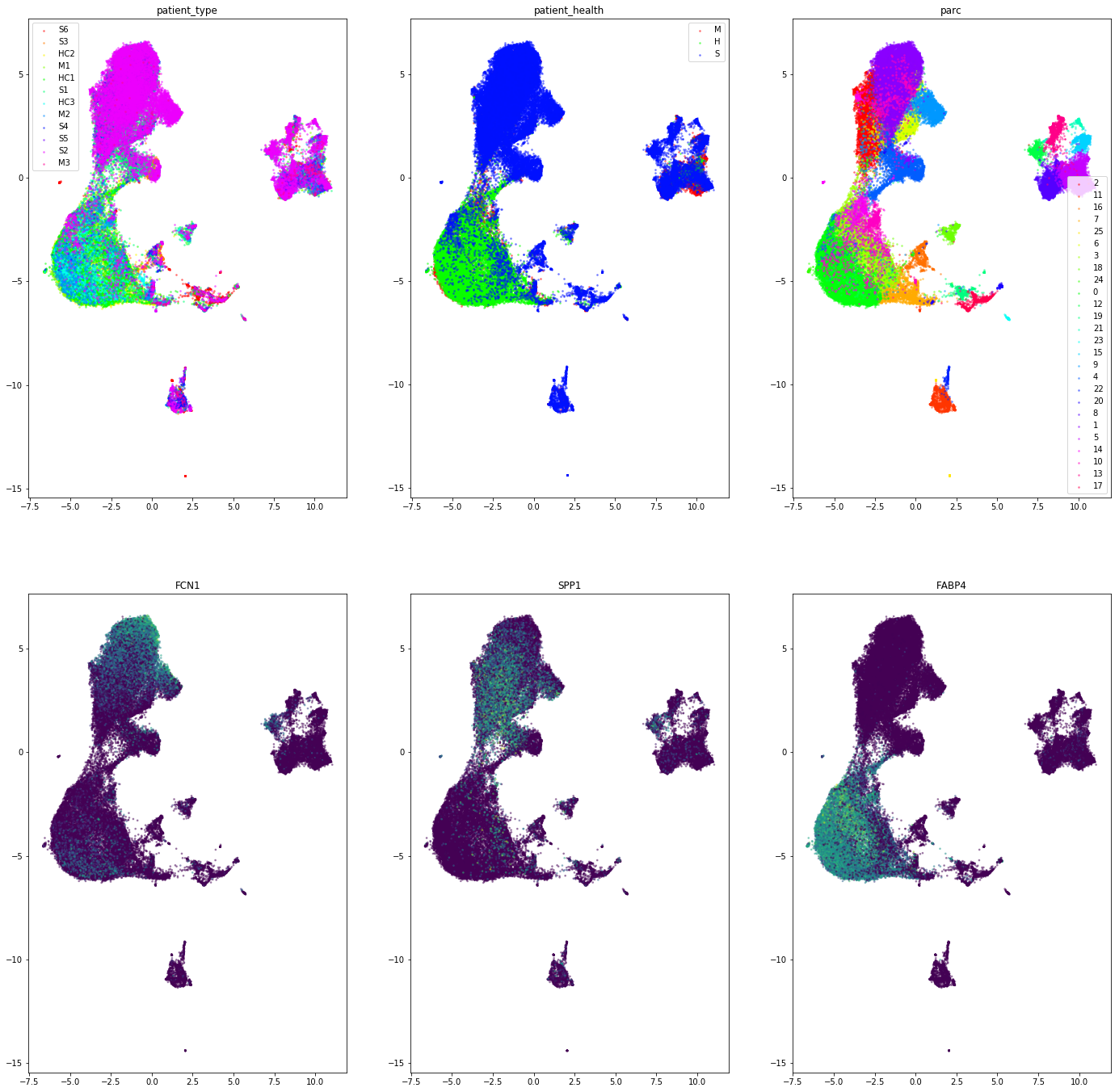
Example Usage on Covid-19 scRNA-seq data
Check out the Jupyter Notebook for how to pre-process and PARC cluster the new Covid-19 BALF dataset by Liao et. al 2020. We also show how to integrate UMAP with HNSW such that the embedding in UMAP is constructed using the HNSW graph built in PARC, enabling a very fast and memory efficient viusalization (particularly noticeable when number cells > 1 Million)
PARC Cluster-level average gene expression

Citing If you find this code useful in your work, please consider citing this paper PARC:ultrafast and accurate clustering of phenotypic data of millions of single cells